How to perform quick search
To facilitate searching on the database, we provide four quick search modes on metagenomic datasets located http://tcm.zju.edu.cn/mgb/advanced.html:
1. search for metagenomic data
2. search for genes of microbiota
3. search for microbiota
4. search for functions of microbiota
Search for metagenomic data
When "General" is selected in the interface (section II), the search will be against metagenomic data.

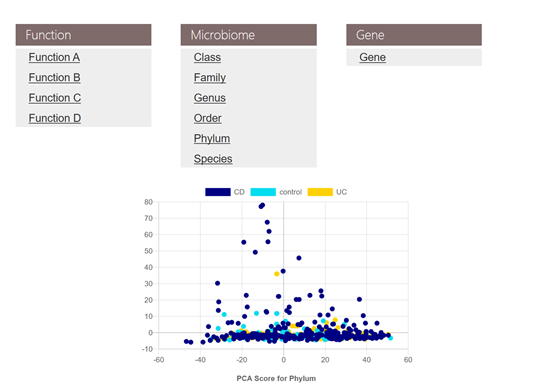
Users are able to search for metagenomic data by any of the listed attributes (section I) in the above figure, e.g. a user is interested to get metagenomic data associated with inflammatory bowel disease (IBD), can select Inflammatory Bowel Disease in the Disease space and press search button. The result of the above search will list the abundance tables for genes, microbiota (phylum, class, order, family, genus, species), functions (hierarchies A, B, C, D), as well as the metadata meeting the searching condition. All above tables are ready for download. Moreover, a PCA score plot for microbiota in phylum level meeting the searching condition is also demonstrated.
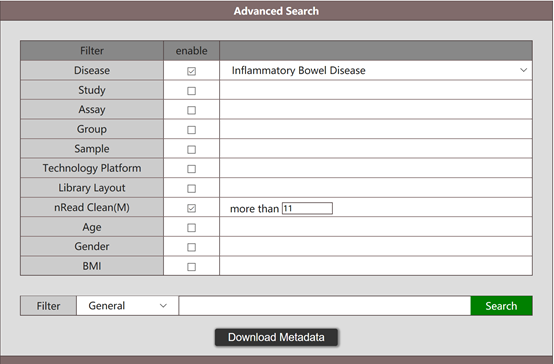
The search is not limited to one search argument, instead the user can combine the search, e.g. search for metagenomic data associated with male population and inflammatory bowel disease.
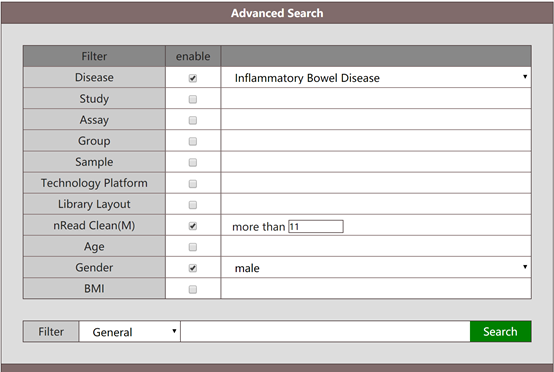
The result of the above search will give a list of all metagenomic data associated with male populations marked with IBD, as well as corresponding metadata.
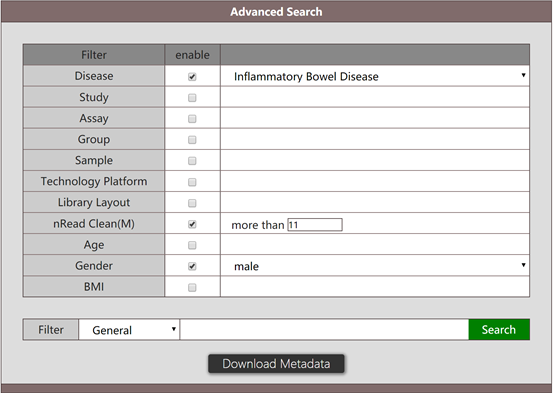
Search for genes of microbiota
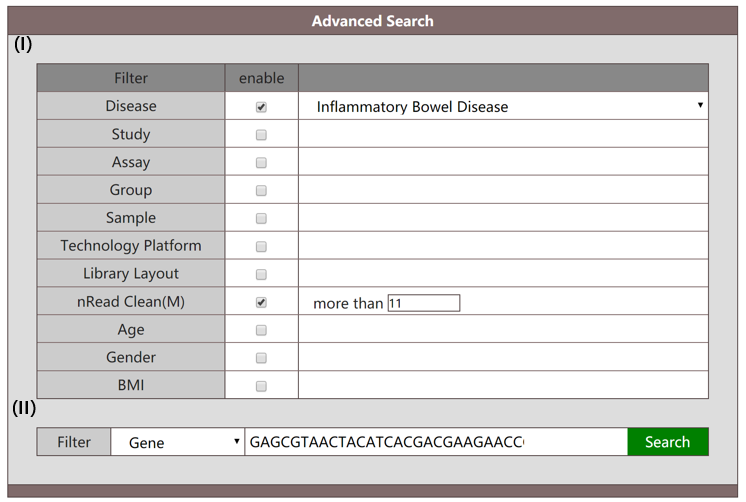
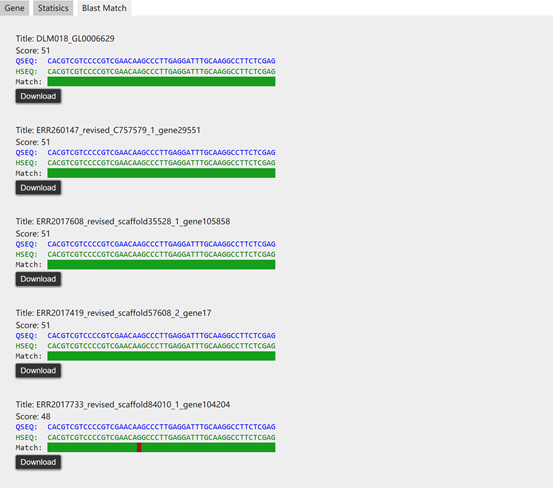
When "Gene" is selected in the interface (section II), the search will be against genes of microbiota. In this mode, the sequence of query gene is also required. The ‘Gene’ mode enables users to search for genes by sequences throughout the whole database, while the attributes provided in section I can be restricted at the same time. The information of target genes in the database matching the query sequence "GAGCGTAACTACATCACGACGAAGAACCGCCGTAACACGCCGGAT" in populations marked with IBD can be retrieved as follows.
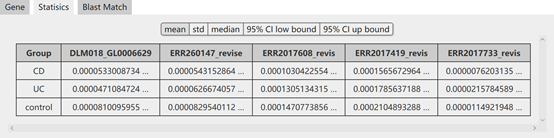
The result of the above search will list the alignment information of top 5 matches and the statistics of them including mean, standard deviation, median and 95% confidence intervals (CIs). Moreover, the abundance of target genes (top 5) is also provided for downlod.

Search for microbiota
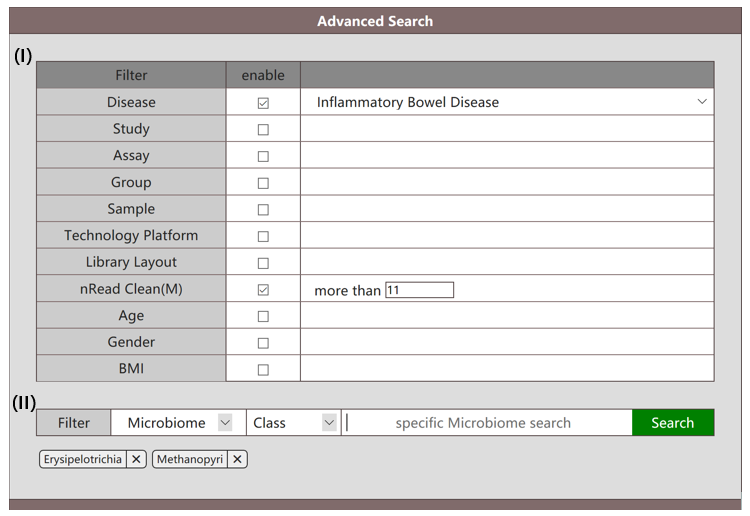
When "Microbiome" is selected in the interface (section II), the "Microbiome" mode enables users to search for microbiota by their names throughout the whole database. Other than the descriptive attributes provided in section I, users need to specify the level (phylum, class, order, family, genus species) of query microbiota, and fill in the names of them in the blank. When users fill in the query microbiome, a list of candidate microbiota are illustrated. Users can select from the drop-down menu for what they want. The selected microbiota will be demonstrated in the bottom left corner of the search interface. The reference files for microbiota in levels phylum, order, class, family, genus, species are provided in Datasets/Supplemental Materials. Names used for search should be selected from the reference files. The information of target microbiota in the database matching the query name "Erysipelotrichia" and "Methanopyri" (class) in populations marked with IBD can be retrieved as follows.
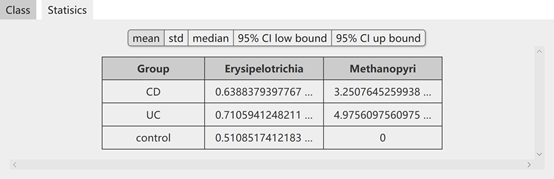
The result of the above search will list the statistics of target microbiota including mean, standard deviation, median and 95% CIs. Moreover, the abundance of target microbiota is also provided for download.

Search for functions of microbiota
When "Function" is selected in the interface (section II), this mode enables users to search for functions by their names throughout the whole database. Other than the descriptive attributes provided in section I, users need to specify the hierarchy (A, B, C, D) of query functions, and fill in the names of them in the blank. When users fill in the query functions, a list of candidate functions are illustrated. Users can select from the drop-down menu for what they want. The reference files for functions in hierarchies A, B, C, D are provided in Datasets/Supplemental Materials. Names used for search should be selected from the reference files. The information of target functions in the database matching the query name "Environmental Information Processing" (A) and "Brite Hierarchies” in populations marked with IBD can be retrieved as follows.
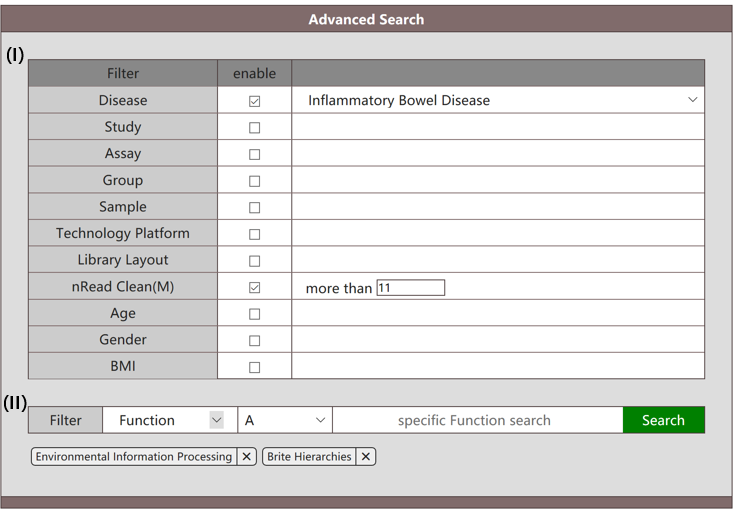
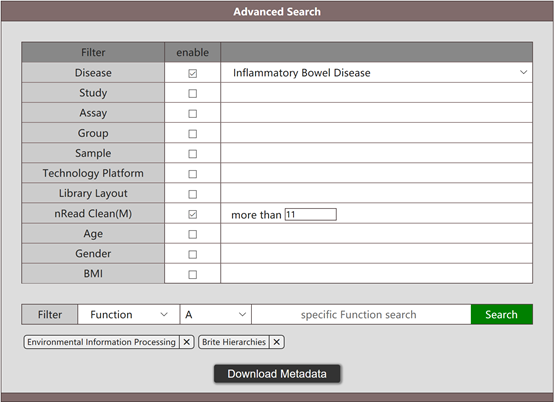
The result of the above search will list the statistics of target functions including mean, standard deviation, median and 95% CIs. Moreover, the abundance table of target functions is also provided for download.

Contact information
College of Pharmaceutical Sciences, Zhejiang University
HangZhou, Zhejiang P.R., China
Tel: (86-571)88208596
Email: fanxh@zju.edu.cn