基于深度学习的单细胞注释工具scDeepSort发布
scDeepSort is a reference-free cell-type annotation tool for scRNA-seq using Deep Learning with a Weighted Graph Neural Network.
基于深度学习的单细胞注释工具scDeepSort已发布在Github和BioRxiv. 第一作者:邵鑫
scDeepSort is a reference-free cell-type annotation tool for scRNA-seq using Deep Learning with a Weighted Graph Neural Network.
Github下载网址:https://github.com/ZJUFanLab/scDeepSort
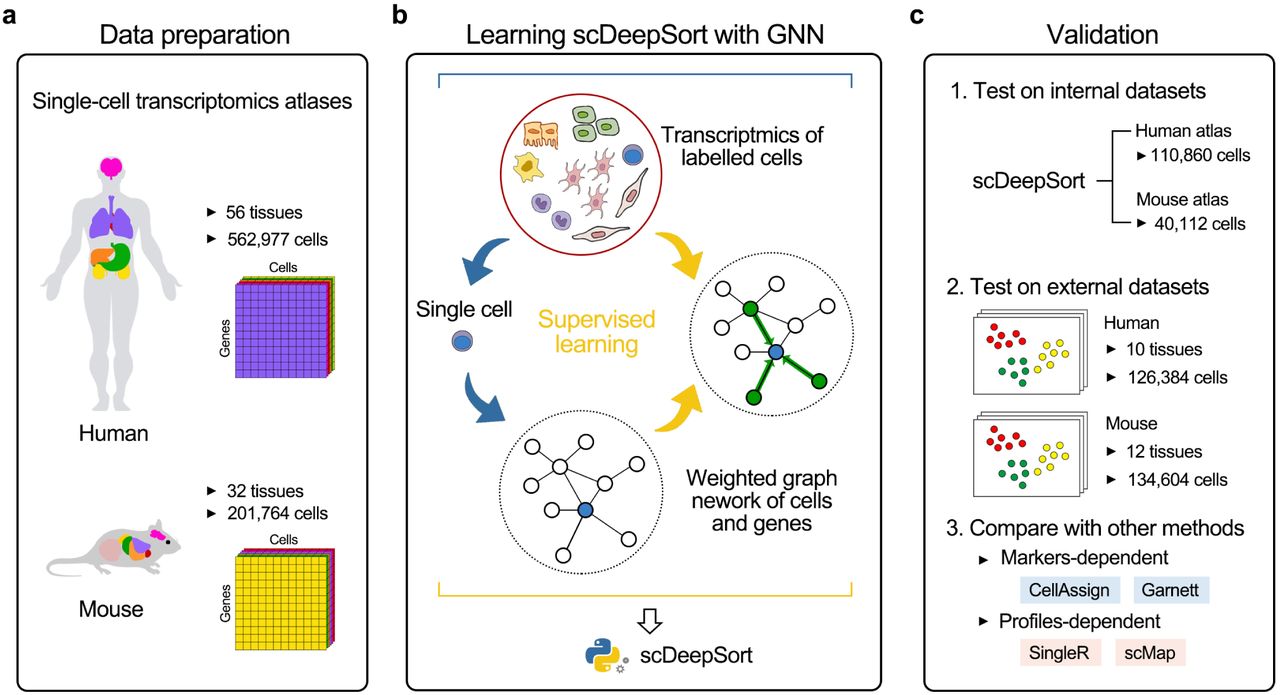
Recent advance in single-cell RNA sequencing (scRNA-seq) has enabled large-scale transcriptional characterization of thousands of cells in multiple complex tissues, in which accurate cell type identification becomes the prerequisite and vital step for scRNA-seq studies.
To addresses this challenge, we developed a reference-free cell-type annotation method, namely scDeepSort, using a state-of-the-art deep learning algorithm, i.e. a modified graph neural network (GNN) model. It’s the first time that GNN is introduced into scRNA-seq studies and demonstrate its ground-breaking performances in this application scenario. In brief, scDeepSort was constructed based on our weighted GNN framework and was then learned in two embedded high-quality scRNA-seq atlases containing 764,741 cells across 88 tissues of human and mouse, which are the most comprehensive multiple-organs scRNA-seq data resources to date. For more information, please refer to a preprint in bioRxiv.

